In order to understand some of the choices we made in our design, it is therefore necessary to consider the biological inspiration of our project. This section aims at providing the required background by presenting an overview of the Embryonics project and of biological inspiration (section 2.1) and then narrowing the focus on the more specific features of bio-inspired hardware (section 2.2).
The analogy between biology and electronics is not as farfetched as it might appear at a first glance. Aside from the more immediate parallel between the human brain and the computer, which has led to the development of fields such as artificial intelligence and is the inspiration for many applications (such as, for example, pattern recognition), a certain degree of similarity exists between the genome(the hereditary information of an organism) and a computer program.
The genome consists of a uni-dimensional string of data encoded in a base-4 system. The DNA (Deoxyribonucleic Acid), the macromolecule in which the genome is encoded, is a sequence of four bases: A (Adenine), C (Cytosine), G (Guanine), and T (Thymine). The information stored on the DNA is chemically decoded and interpreted to determine the function of a cell. A computer program is a uni-dimensional string of data encoded in a base-2 system (0 and 1). Stored in an electronic memory circuit, it is interpreted to determine the function of a processor.
Of course, carbon-based biology and silicon-based computing are different enough that no straightforward one-to-one relationship between the genome and a computer program (and indeed between any biological and computing process) can be established, except at a very superficial level. However, through careful interpretation, some basic biological concepts can be adapted to the design of computer systems, and some biological processes are indeed extremely interesting from a computer designer's point of view: for example, an organism's robustness (achieved by its healing processes) is unequaled in electronic circuits, while the natural process of evolution has produced organisms of a complexity which far exceeds that of modern computer systems.
It should be obvious that, biology being a very vast field, the biological concepts which could potentially be adapted to computer hardware are numerous, as are the approaches to applying such concepts to electronics. The first step in such an effort should therefore be to identify which aspects of the biological world will be studied, and define a unified framework which can meld them into a single project.
Even if the first direction is, so far, definitely dominant (computers are quickly becoming indispensable tools for medical and biological applications ranging from computerized axial tomography to genomic sequence analysis), biology is starting to inspire engineers to develop quasi-biological systems. Among the best-known examples of such systems, we can mention:
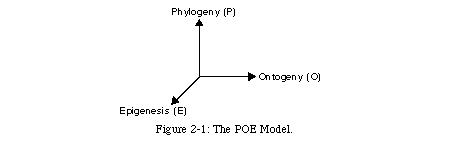
Obviously, biological inspiration in computer science can assume a variety of different forms, and Embryonics does not attempt to cover all its possible aspects. Nevertheless, the scope of the project is such that it does involve many of the traditional bio-inspired systems, as well as the development of novel approaches. To illustrate the scope of the project, we have created the POE model [84, 90], an attempt to classify bio-inspired systems, according to their biological source of inspiration, along three axes: phylogeny, ontogeny, and epigenesis (Fig. 2-1).

On the phylogenetic axis we find systems inspired by the processes involved in the evolution of a species through time, i.e. the evolution of the genome. The process of evolution is based on alterations to the genetic information of a species, occurring through two basic mechanisms: crossover and mutation.
Crossover (or recombination) is directly related to the process of sexual reproduction. When two organisms of the same species reproduce, the offspring contains genetic material coming from both parents, and thus becomes a unique individual, different from either parent. Mutation consists of random alterations to the genome caused either by external phenomena (i.e., radiation or cosmic rays) or by chemical faults which occur when the two genomes merge. Often resulting in non-viable offspring2, mutations are nevertheless vital for the process of evolution, as they allow "leaps" in evolution which would be impossible to achieve by merging the genomes of individuals of the same species.
Both these mechanisms are, by nature, non-deterministic. This represents both their strength and their weakness, when applied to the world of electronics. It is a strength, because they are fundamentally different from traditional algorithms and thus are potentially capable of solving problem which are intractable by deterministic approaches. It is a weakness, because computers are inherently deterministic (it is very difficult, for example, to generate a truly random number, a basic requirement for non-deterministic computation, in a computer).
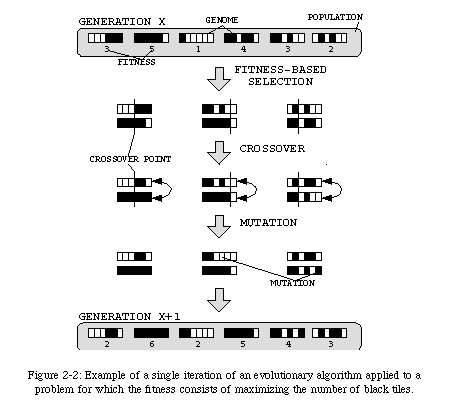
Even with this disadvantage, algorithms which exploit phylogenetic mechanisms are carving themselves a niche in the world of computing. These algorithms, commonly referred to as evolutionary algorithms(Fig. 2-2) (a label which regroups domains such as genetic algorithms [65], evolutionary programming [28], and genetic programming [48, 49]), are usually applied to problems which are either too ill-defined or intractable by deterministic approaches, and whose solutions can be represented as a finite string of symbols (which thus becomes the equivalent of the biological genome). An initial, random population of individuals (i.e., of genomes), each representing a possible solution to the problem, is iteratively "evolved" through the application of mutation (i.e., random alterations of a sequence) and crossover (i.e., random mergings of two sequences). The resulting sequences are then evaluated on the basis of their efficiency in solving the given problem (the fitness function) and the best (fittest) solutions are in turn evolved. This approach is not guaranteed to find the best possible solution to a given problem, but can often find an "acceptable" solution more efficiently than deterministic approaches. Our laboratory is very active in this domain [86, 87, 88].
It appears, then, that the phylogenetic axis has already provided a considerable amount of inspiration to the development of computer systems. To date, however, its impact has been felt mostly in the development of software algorithms, and only marginally in the conception of digital hardware. Koza et al. pioneered the attempt to apply evolutionary strategies to the synthesis of electronic circuits when they applied genetic algorithms to the evolution of a three-variable multiplexer and of a two-bit adder [50]. Also, evolutionary strategies have been applied to the development of the control circuits for autonomous robots [26, 27]. Unfortunately, technical issues have posed severe obstacles to the progress of this kind of approach, and few groups are currently active in this domain [37, 38, 100]. Our laboratory contributed to the research with the development of Firefly [33], a machine capable of evolving a solution to the problem of synchronizing a uni-dimensional cellular automaton (see the next chapter for a more detailed description of cellular automata). This relatively simple task represents nevertheless the first example of hardware capable of evolving completely on-line, i.e. without the assistance of a computer.
The human genome contains approximately 3x109bases. An adult human body contains something like 6x1013cells, of which approximately 1010 are neurons, with 1014 connections. Obviously, the genome cannot contain enough information to completely describe all the cells and synaptic connections of an adult organism [21].
There must therefore exist a process which allows the organism to increase in complexity as it develops. Since we already know that the additional information cannot come from within the cell itself, the only possible source of additional information is the outside world: the growth and development of an organism must be influenced by the environment, a process which is most evident in the development of the nervous, immune, and endocrine systems. Following A. Danchin's terminology [21, 22], we have labeled this process epigenesis3.
Epigenetic mechanisms have already had considerable impact on computer science, and particularly on software design, notably through the concept of learning. The parallel between a computer and a human brain dates to the very earliest days of the development of computing machines, and led to the development of the field known as artificial intelligence[108].
AI, which probably knew its greatest popularity in the seventies, tried to imitate the high-level processes of the human mind using heuristic approaches. After a number of years in the spotlight, this field seems today to have lost momentum, but not without leading to useful applications (such as expert systems) and major contributions to the development of novel algorithms (for example, IBM's well-known Deep Blue computer exploits algorithms largely derived from AI research).
Hardware systems based on epigenetic processes are starting to emerge, in the form of artificial neural networks (ANNs) [9, 36], two-dimensional arrays of processing elements (the neural cells) interconnected in a relatively complex pattern (Fig. 2-3). Each cell receives a set of input signals from its neighbors, processes them according to a given, implementation-dependent function, and propagates the results through the network. This process is a good approximation of the mechanisms exploited by biological neurons (for example, as in nature, some inputs signals have a greater impact, or weight, in the computation of a neuron's output value).
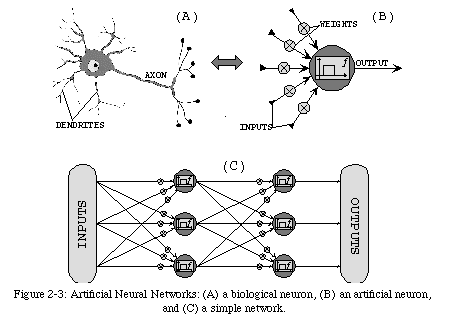
ANNs cannot be expected to perform correctly (i.e. to produce the correct output) for allinput patterns, a drawback which prevents their use in many applications. However, they have proved their worth (to the point that they are starting to be adopted in commercial systems) in applications such as voice and character recognition, where a limited margin of error is acceptable.
Our laboratory is also involved in research along the epigenetic axis, with the development of FAST (Flexible Adaptable-Size Topology) [74, 75, 76], a neural network with a dynamically reconfigurable structure. Traditional ANNs have a fixed interconnection structure, and only the weights associated with the connections can be modified. By implementing the network using reconfigurable logic (see subsection 2.2.2 ), FAST achieves on-line learning capabilities and can exploit an adaptable topology (two features which are essential to obtain true learning systems, as opposed to "learned" ones). While FAST is not the first neural network based on an adaptable topology [30, 67], it is unique in that it does not require intensive computation to reconfigure its structure, and can thus exist as a stand-alone machine which could be used, for example, in real-time control applications.
The phylogenetic and epigenetic axes of the POE model cover the great majority of existing bio-inspired systems. The development of a multi-cellular biological organism, however, involves a set of processes which do not belong to either of these two axes. These processes correspond to the growth of the organism, i.e. to the development of an organism from a mother cell (the zygote) to the adult phase. The zygote divides, each offspring containing a copy of the genome ( cellular division ). This process continues (each new cell divides, creating new offspring, and so on), and each newly formed cell acquires a functionality (i.e., liver cell, epidermal cell, etc.) depending on its surroundings, i.e., its position in relation to its neighbors ( cellular differentiation ).
Cellular division is therefore a key mechanism in the growth of multi-cellular organisms, impressive examples of massively parallel systems: the 6x10 13cells of a human body, each a relatively simple elements, work in parallel to accomplish extremely complex tasks (the most outstanding being, of course, intelligence). If we consider the difficulty of programming parallel computers (a difficulty which has led to a decline in the popularity of such systems), biological inspiration could provide some relevant insights on how to handle massive parallelism in silicon.
A fundamental feature of biological organisms is that each cell contains the blueprint for the entire organism (the genome), and thus can potentially assume the functionality of any other cell: no single cell is indispensable to the organism as a whole. In fact, cells are ceaselessly being created and destroyed in an organism, a mechanism at the base of one of the most interesting properties of multi-cellular organisms: healing.
The mechanisms of ontogeny (cellular division and cellular differentiation), unlike those of epigenesis and phylogeny, are completely deterministic, and are thus, in theory, more easily adaptable to the world of digital circuits (which is by nature deterministic). In spite of this, the ontogenetic axis has been almost completely ignored by computer scientists, despite a promising start in the fifties with the work of John von Neumann, who developed a theoretical model of a u niversal constructor, a machine capable of constructing any other machine, given its description [104]. Given a description of itself, the universal constructor can then self-replicate, a process somewhat analogous to cellular division.
We will describe von Neumann's machine, one of the fundamental sources of inspiration for the Embryonics project in general and for this work in particular, in detail in the next chapter, but we should mention that, unfortunately, electronic circuits in the 1950s were too primitive to allow von Neumann's machine to be realized, and the concept of self-replicating machines was thus set aside.
Probably the main obstacle to the development of self-replicating machines was the impossibility of physically creating self-replicating hardware. In fact, such machines require a means to transforming information (i.e. the description of a machine) into hardware, and such a means was definitely unpractical until recently. As we will see below, the introduction of programmable logic circuits, by demonstrating the feasibility of such a process, was an important step towards the development of self-replicating machines. One of the main goals of the Embryonics project is to determine if, given modern technology, Von Neumann's dream of a self-replicating machine can be realized in hardware. The work presented in this thesis represents a major step in this direction: by creating a hardware system capable of self-replication, we open the way to the practical realization of von Neumann's universal constructor.
Cell, n.: the basic structural and functional unit of all organisms; cells may exist as independent units of life (as in monads) or may form colonies or tissues as in higher plants and animals.
Nucleus, n.: a part of the cell containing DNA and RNA and responsible for growth and reproduction.
DNA, n.: a nucleic acid consisting of large molecules shaped like a double helix; associated with the transmission of genetic information.
RNA, n.: a nucleic acid that transmits genetic information from DNA to the cytoplasm; controls certain chemical processes in the cell.
In other terms, the cell is the smallest structural unit of an organism which contains the description of the entire organism, i.e., its genome, which is interpreted to control the cell's functions. One approach which allows us to respect these definitions is to implement computing systems (the artificial organisms) using an array of (relatively) small processing elements (the artificial cells), each executing the same program (the artificial genome). As we will see, this approach allows us not only to respect the fundamental definitions of a biological cell, but also to exploit some of the more specialized mechanisms on which the ontogenetic development of a cell is based.Genome, n: one haploid set of chromosomes with the genes they contain; the full DNA sequence of an organism.
Many of the precise mechanisms whereby the genome is accessed and interpreted inside each cell are still unknown to science5. Since our intention is notto imitate biology, however, such detailed knowledge is superfluous: the transition from the carbon-based world of biology to the silicon-based world of electronic circuits will necessarily generate considerable discrepancies. Undoubtedly the greatest such discrepancy is a consequence of the process of biological ontogeny itself, which is based on biology's capability to handle physical material: biological ontogenetic development is based on the physicalgrowth and replication of cells, and biological self-repair is a consequence of their physicalcreation and destruction. The state of the art in the design of electronic circuits does not allow us to act on the physical structure of our cells (i.e., the silicon) after fabrication.
This obstacle, which was a major factor in preventing the realization of von Neumann's universal constructor, would have been almost insurmountable until a few years ago, when the first FPGA circuits were developed.
FPGAs are quickly becoming an essential tool for the design of complex computer circuits. In the industry, they are mainly used for rapid prototyping : manufacturing a VLSI circuit is an expensive and time-consuming endeavor, and a circuit has to be tested very extensively before it is sent to a foundry for fabrication. Most design errors can be detected through software simulation, but the speed (or lack thereof) of such simulations does not allow extensive testing of complex circuits. By using FPGAs to implement a design, the circuit can be tested at hardware speeds, and the debugging can therefore be much faster and more complete (moreover, some faults can only be observed when the circuit is tested at speed).
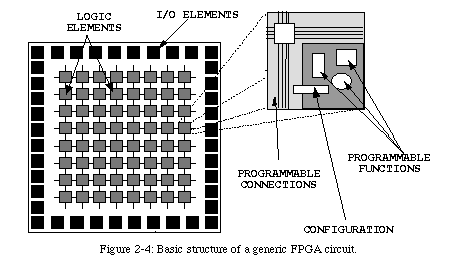
However, in some cases the versatility of FPGAs can overcome this shortcoming, either because the circuit is not speed-critical, but could benefit from regular upgrades or even dynamic alterations, or because the advantage of having dedicated (often parallel) processors can easily compensate the slower clock rate (for example, specific mathematical operations which would require many clock cycles in a general-purpose processor, but could be executed in a single, if slower, clock cycle in a dedicated processor). Given the prohibitive cost of developing a dedicated computer system in VLSI, using programmable logic can be a viable and cost-effective option in many applications.
Our laboratory has been involved in this kind of FPGA-based systems for many years. Among the most interesting project, we can mention:
Since the Embryonics project is not meant to produce a commercially viable product8, speed of operation is not one of our main priorities. On the other hand, the reprogrammability of FPGAs is a solution to the problem of implementing an ontogenetic machine, as it provides a way to modify the hardware structure of a system by altering information, sidestepping the need to handle physical matter.
Drawing inspiration from biological organisms has thus led to define our organism as a two-dimensional array of processing elements, all identical in structure (each cell must be able to execute any subset of the genome program) and each executing a different part of the same program, depending on its position. At first glance, this kind of system might not seem very efficient from the standpoint of conventional circuit design: storing a copy of the genome program in each processor is redundant, since each processor will only execute a subset.
However, by accepting the weaknesses of bio-inspiration, we can also partake of its strengths. One of the most interesting features of biological organisms is their robustness, a consequence of the same redundancy which we find wasteful: since each cell stores a copy of the entire genome, it can theoretically replace any other. Thus, if one or more cells should die as a consequence of a trauma (such as a wound), they can be recreated starting from any other cell. By analogy, if one or more of our processors should "die" (as a consequence, for example, of an hardware fault), they can theoretically be replaced by any other processor in the array.
The redundancy introduced by having multiple copies of the same program thus provides an intrinsic support for self-repair, one of the main objectives of our research: by providing a set of spare cells (i.e., cells that are inactive during normal operation, but which are identical to all other cells and contain the same genome program), we are able (Fig. 2-5) to reconfigure the array around one or more faulty processors (of course, as in living beings, too many dead cells will result in the death of the entire organism).
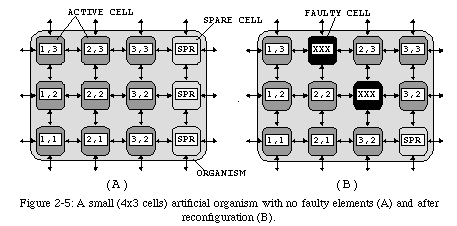
Moreover, if the function of a cell depends on its coordinates, the task of self-replication is greatly simplified: by allowing our coordinates to cycle (Fig. 2-6) we can obtain multiple copies of an organism with a single copy of the program (provided, of course, that enough processors are available). Depending on the application and on the requirements of the user, this feature can be useful either by providing increased performance (multiple organisms processing different data in parallel) or by introducing an additional level of robustness (the outputs of multiple organisms processing the same data can be compared to detect errors).
While the structure of the functional unit can vary, biological organisms provide a powerful example of complex behavior derived not from the complexity of the components, but from the parallel operation of many simple elements. One of the goals of our project is to show that biological inspiration allows us, without excessive difficulty, to design complex systems by combining very simple cells.
To illustrate the capabilities of our system, we have realized in hardware a first prototype of such a cell, known as MicTree(for tree of microinstructions) [58, 59, 60, 93], where the functional unit is a 4-bit register, the coordinate registers are 4 bits wide (limiting the maximum size of organism to 16x16 cells, sufficient for demonstration purposes, but restrictive for real applications), and communications, implemented using directional 4-bit busses, are limited to the cardinal neighbors. To write genome programs for MicTree, we have developed a small (but complete) language, complex enough to potentially implement any application, but simple enough that the interpreter need not be large. The instructions of the language are as follows:
where REGis the 4-bit register in the functional unit, Xand Yare the two 4-bit coordinates, MASK lets the bits of REGbe accessed individually, and VAROUT and VARINidentify one of the four possible I/O busses. The instructions are coded in 8-bit words, and the genome memory, of fixed size, can store up to 1024 such words. Each MicTree cell, realized using an Actel A1020B [3] FPGA (ideally suited for prototyping), was then mounted on a custom printed circuit board with a set of 7-segment displays and LEDs (Light Emitting Diodes), which in turn was inserted in an 8X8cm plastic box, the Biodule 601 (Fig. 2-9). These boxes can be fitted together, like a jigsaw puzzle, to form a two-dimensional array of cells and provide a set of neighbor-to-neighbor connections without additional cables.
These applications, because of the limited size of our prototype, are relatively simple. Nevertheless, they are interesting in that they exhibit both the properties of self-repair (provided spare cells are available) and self-replication (if the array is large enough to contain multiple copies of the organism). In designing applications for our prototype, however, we came to realize that, while MicTree contains all of the minimal features required by our electronic cell, its fixed structure seriously limits the range of applications it can efficiently implement.
One of the most constraining factors is probably the functional unit: a single 4-bit register is too small for complex applications. Of course, the functional unit is not, by itself, constraining, as it could be compensated by using either a larger organism (cells can be very simple, if many are working in parallel) or a longer genome program. However, since both the scope of the coordinates and the size of the genome memory are also fixed, MicTree is unsuited for complex applications.
Therefore, as we suspected, we need to able to reprogram the hardware itself to create a truly ontogenetic machine. The features of the functional unit (e.g., the size of the data path) have to fit the application to reduce the complexity on the rest of the cell. The genome can have very different sizes depending on the complexity of the task (there usually is a trade-off between hardware and software, that is between the complexity of the functional unit and the length of the genome), and the size of the genome memory should therefore be programmable, a feature has an impact on the complexity of the interpreter. The size of the registers containing the coordinates depends on the size of the organism. A particular application might be more efficiently executed by a specific connection pattern. Obviously, we can design a complex enough cell that it will satisfy the greatest majority of possible applications. In fact, with a complex enough interpreter (and thus genome language) or functional part, we can be limited only by size of the genome memory. In practice, however, this solution would be very wasteful: no benefit could be achieved through tailoring processor to application, and the processors would become so complex that we would reintroduce all the drawbacks which have kept conventional multiprocessor parallel systems from gaining widespread acceptance (such as the difficulty of writing parallel code and the complexity of handling communication between processors).
Once again, biology provides a possible solution: the physical structure of a biological cell is determined by chemical processes occurring at the molecular level. Having introduced the concepts of electronic organism and of electronic cell, we now need to define the concept of electronic molecule. Fortunately, we are familiar with a kind of circuit which can be used to implement our molecular layer: FPGAs. Using programmable gate arrays as our molecules allows us to maintain the fundamental analogy with the world of biology: whereas a living cell consists of a three-dimensional array of molecules, a processor consists of a two-dimensional array of logic gates. Further, since in biology the most complex mechanisms of the cellular layer (notably, the genome program and the coordinate system) do not concern the molecular layer, the structure of our electronic molecule is not unlike that of conventional FPGA logic elements. We are therefore confronted with a three-layer system (Fig. 2-10), summarized in Table 2-1.
| Biology | Electronics |
|---|---|
| Multi-cellular organism | Parallel computer system |
| Cell | Processor |
| Molecule | FPGA element |
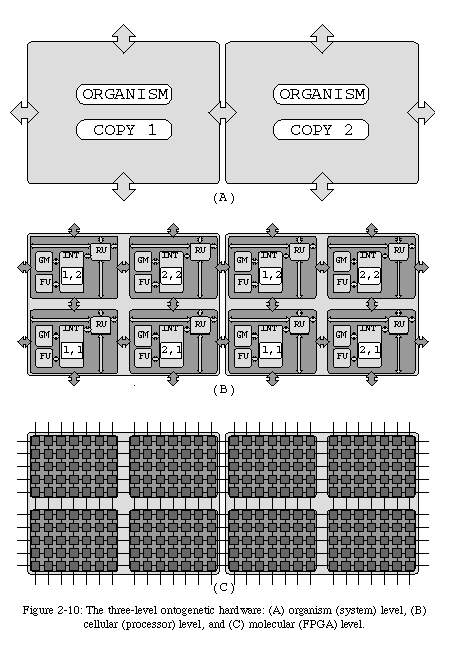
The first such feature concerns the basic structure of our artificial organism: a two-dimensional array of identical processing elements (cells). The dimensions of such an array can be very substantial, since a processor, however simple, is nevertheless a relatively complex circuit, and restricting the amount of programmable logic to a single chip can quickly become a serious constraint. Envisaging the use of more than one chip, however, implies the presence of a mechanism allowing designs to be easily distributed among multiple chips. Three main factors inhibit this process in conventional FPGAs:
The second feature of ontogenetic hardware we need to implement is healing, i.e., in a terminology more familiar to computer designers, self-repair.
As we mentioned, the cellular level, through its coordinate system, provides an intrinsic capability for self-repair. However, while in biological systems the death of a cell is a commonplace occurrence, in our electronic organism we can not afford the death of many cells: if we assume that a cell will require something in the order of a few hundred molecules, and we assume a finite number of available molecules, we obviously need to minimize the loss of cells. As a consequence, we need a mechanism to repair minor faults at the molecular level, which in turn requires a mechanism which allows self-test, i.e. the detection of faults. In fact, a prerequisite for being able to repair faults, both at the cellular and at the molecular level, is the capability of detecting the occurrence of such faults. Moreover, if the self-repair mechanism is to operate on-line (i.e., while the application is running, rather than in a separate phase), self-test must also be performed on-line.
The design of such a mechanism is a considerable challenge, particularly since the size of the FPGAs elements is such that the additional logic required by the process be minimized. However, the absence of molecular-level self-test would imply that the detection of faults be handled at the cellular level, and since the cells are application-dependent, the self-test logic would have to be included in every new cell, which would complicate their design to a considerable degree.
Providing an on-line self-test and self-repair mechanism at the molecular level is therefore, if not required, at least very desirable, since it would not only increase the robustness of the system (which would "survive" a larger number of faults), but also substantially simplify the design of the cells (by moving much of the additional logic to the application-independent molecular level). Fortunately, the task of designing such a mechanism is somewhat simplified by the presence of a second level of robustness at the cellular level, which implies that the molecular-level self-repair need not be able to handle all faults occurring in the circuit.
To efficiently implement our electronic organism, we would therefore require a completely homogenous self-repairing FPGA which can easily be configured as an array of identical processing elements (self-replication). No commercial FPGA provides the features we need, and it is doubtful whether the demand for self-repair and self-replication is sufficient to justify the additional logic in a commercial circuit, at least in the foreseeable future. As consequence, the efficient implementation of ontogenetic hardware requires the conception of a new FPGA, capable of self-replication and self-repair. The development of such an FPGA constitutes the main challenge of the ontogenetic axis of the Embryonics project, and is the focus of this thesis.
2.In fact, the great majority of mutations have no effect, as they act on unused parts of the genome.
3.This label is by no means universally accepted. We thank Prof. Marcello Barbieri of the University of Ferrara, Italy, for bringing this fact to our attention.
4.Of course, considering the different environments, the analogy will necessarily be limited to the most macroscopic features.
5.It is our secret hope that, in some indirect way, our work might be of some use to biologists by suggesting possible mechanisms for accessing and decooding the biological genome.
6.In fact, some FPGAs can implement analog circuits as well.
7.The elements of an FPGA are usually referred to as cells, but be will avoid the term so as not to engender confusion with our electronic cells.
8.At least, not directly: we do hope that some of the strategies and mechanism developed in our project will eventually, duly adapted, have repercussions on mainstream circuit design.
9.When dealing with the world of electronics circuits, the current state of the art in circuit fabrication limits us to two dimensions, but this limitation (being quantitative, rather than qualitative) is not particularly constraining, and should not have serious consequences for our project.
10.This kind of parallel systems are a particular case of what are commonly known as Single-Program Multiple-Data (SPMD) computer systems [5].
11.Complex organisms can have rather complicated mechanisms which allow long-distance communication between cells to occur, but these can be realized using local connections, with a loss of efficiency compensated by the gain in simplicity.