|
 |
||
|
|
|
||
|
|
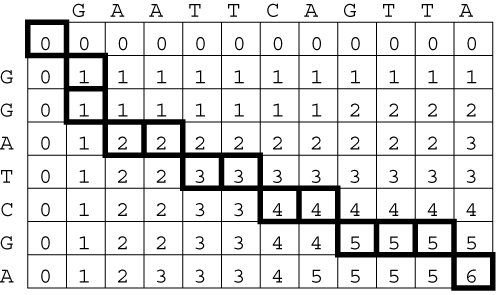
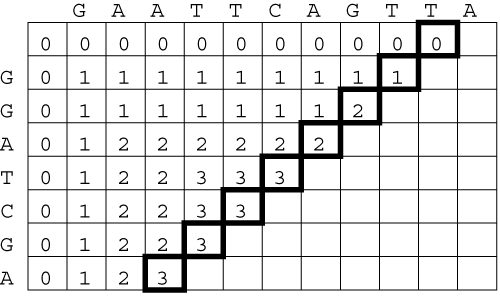
Nowadays the comparison and the alignment of characters taken from a finite alphabet is a fundamental task in many applications, ranging from full-text search to computational biology. In particular, string comparison is a critical issue in the field of molecular biology. In fact, both DNA fragments and proteins can be represented as sequences of characters (taken from alphabets of 4 and 20 symbols, respectively) and sequence similarities provide useful information about the functional, structural and evolutionary relationships between the corresponding molecules. Biological sequences may differ because of local substitutions, insertions and deletions of one or more characters. The complexity of string comparison comes from the large number of possible combinations of these three basic mutations. The similarity between two strings can be evaluated either in terms of edit distance or in terms of similarity score. The edit distance is a measure of the minimum number of mutations that may have transformed the first string in the second, or, in other terms, of the minimum number of edit operations required to make the two strings equal to each other. The similarity score is a measure of the maximum number of residual matches between the two strings. Both metrics can be evaluated in polynomial time by means of dynamic programming techniques.
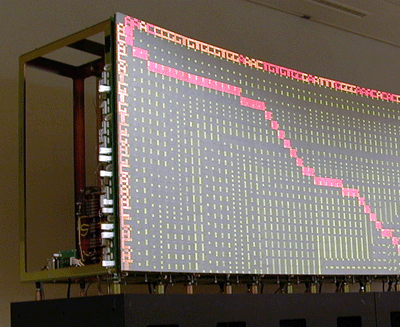
The key algorithm for evaluating the similarity between two strings of length N and M was developed by Needleman and Wunsch and takes O(NxM) steps to complete execution. The structure of the algorithm makes it suitable for a parallel implementation on a systolic array. In particular, hardware parallelism can be exploited to perform string comparison in O(N+M) steps. In this experience we present a parallel implementation of the Needleman-Wunsch algorithm on the BioWall.
The BioWall cannot compete in performance with existing parallel implementations of the Needleman-Wunsch algorithm, since it suffers from the typical performance limitations of a large prototyping platform. Nevertheless, the implementation of the Needleman-Wunsch algorithm on the BioWall is a significant design experiment in the field of reconfigurable computing because of the peculiarities of the target architecture.
For further information
Resources
|